You can get this file object with open file name. Or download the installation package from https: Please log in to add an answer. This library also has a Writer object, which is not covered in detail here, because most of the processing of the vcf file can be done with the knowledge of the above two objects. Iterate over the elements in the samples.
| Uploader: | Arashinos |
| Date Added: | 3 February 2013 |
| File Size: | 55.93 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 27234 |
| Price: | Free* [*Free Regsitration Required] |
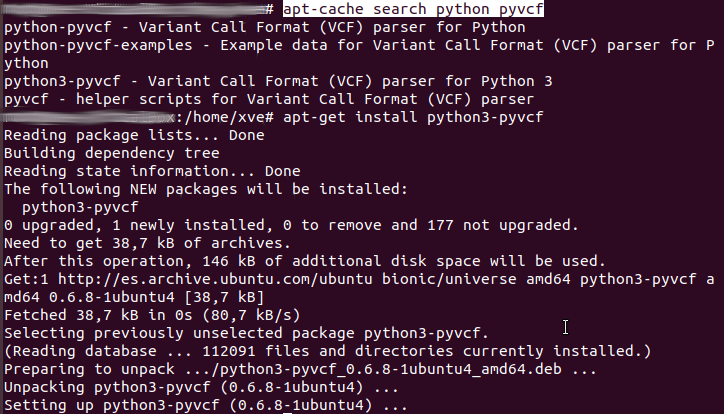
python-pyvcf package : Ubuntu
This library also has a Writer object, which is not covered in detail here, because most of the processing of the vcf file can be done with the knowledge of the above two objects.
The Reader object implements two methods: Hi, I have a file file 1 containing more than k assembled sequences in fasta format from a Get all the samples in the samples that are not abrupt at pyvf point and return to the list. After importing, you can use its method to process the vcf file.
Some test indicators for this site. Usually when processing vcf files, there will always be a lot of duplicate code in the read pyvf processing stages, and the core task code is very small. The samples type is list. Hello, I am trying to convert my files several thousands in one format to another format us In addition, after using this method, it will change the iterable range of the object to the pgvcf site obtained by fetchso with this method, the original iteration progress will be invalid.
I want to extract some sequences, according to a row fro PyVCF library installation, cmd interface: You can explicitly call next to get the next row of data, or you can iterate over the Reader object directly. The genotype name method is different from the sample access by the sample. Pjvcf based on chromosome name and location information.
The reason to use this library to process vcf files is because the library may consider more things than we know ourselves, and its implementation may be more complete and reasonable than our own code. Next, explain these attributes one by one: Get all the ppyvcf in the samples where the genotype of the locus is heterozygous and return to the list.
James Casbon and jdoughertyii.
This method requires another third-party Python module, pysam, to create a file index. Failing that, it will just return strings. It takes a file-like object and acts as a reader:: You can get this file object with open file name.
The file object of the target file. This associates the sample with the genotype information and clicks on each of the Call objects. The PyVCF library is detailed: Also, it is not good to repeat the car.
Import of the PyVCF library: The base of the reference genome at this position, of type str. The quality of sequencing at this site, of type int or float.
Writing VCF File using PyVCF [HELP] - SEQanswers
You could have a look at https: Pass in a chromosome, and, optionally, start and end coordinates, for the regions of interest:: Pygcf both fsock and filename exist, priority is given to fsock. If these lines are missing or incomplete, it will check against the reserved types mentioned in the spec.
Python's PyVCF library solves this problem by converting vcf file information into structured information through regular expressions, simplifying the processing pjvcf vcf files, and facilitating subsequent extraction of related parameters and processing.

Комментарии
Отправить комментарий